Assembly Overview:  TOP
TOP
BLAST on RefSeq (Human):  TOP
TOP
| | Locus | Definition | Score | E value | FL |
|---|
 |
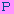 | | zinc finger protein 703 | 30.4 | 1.9 | |
BLAST on RefSeq (Mouse):  TOP
TOP
| | Locus | Definition | Score | E value | FL |
|---|
BLAST on RefSeq (Dog):  TOP
TOP
| | Locus | Definition | Score | E value | FL |
|---|
BLAST on RefSeq (Cattle):  TOP
TOP
| | Locus | Definition | Score | E value | FL |
|---|
BLAST on RefSeq (Pig):  TOP
TOP
| | Locus | Definition | Score | E value | FL |
|---|
Alignment:  TOP
TOP
20060611C-008881 : ............................................................
---------+---------+---------+---------+---------+---------+ 0
SPL01_0053_F09.b : nnnnggcattgtactaagacxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxx
SPL01_0053_F08.b : nnnttgctaggactatnacxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxx
20060611C-008881 : ......................GCTTTTATGATTTAACGTTACTGTGTAAGTACTTTCCC
---------+---------+---------+---------+---------+---------+ 38
SPL01_0053_F09.b : xxxxxxxxxxxxxxxxxxxxxxGCTTTTATGATTTAACGTTACTGTGTAAGTACTTTCCC
SPL01_0053_F08.b : xxxxxxxxxxxxxxxxxxxxxxGCTTTTATGATTTAACGTTACTGTGTAAGTACTTTCCC
20060611C-008881 : ACATTGCTACACTTGCTTGGTGGTTCTCATGGAAATTCCATTTTCCATGATCTTGACATT
---------+---------+---------+---------+---------+---------+ 98
SPL01_0053_F09.b : ACATTGCTACACTTGCTTGGTGGTTCTCATGGAAATTCCATTTTCCATGATCTTGACATT
SPL01_0053_F08.b : ACATTGCTACACTTGCTTGGTGGTTCTCATGGAAATTCCATTTTCCATGATCTTGACATT
20060611C-008881 : CTGGCTGGTGTTTCTGTCATCGTCGACTCTGGCTTTCTGTTACCTGAGTCAGGGTGTCTC
---------+---------+---------+---------+---------+---------+ 158
SPL01_0053_F09.b : CTGGCTGGTGTTTCTGTCATCGTCGACTCTGGCTTTCTGTTACCTGAGTCAGGGTGTCTC
SPL01_0053_F08.b : CTGGCTGGTGTTTCTGTCATCGTCGACTCTGGCTTTCTGTTACCTGAGTCAGGGTGTCTC
20060611C-008881 : TCCGGTCTGAGCTCTCGATCACACTTAGCTTTCCTCTTGCATGTCCTGTGTCAAGCAGCT
---------+---------+---------+---------+---------+---------+ 218
SPL01_0053_F09.b : TCCGGTCTGAGCTCTCGATCACACTTAGCTTTCCTCTTGCATGTCCTGTGTCAAGCAGCT
SPL01_0053_F08.b : TCCGGTCTGAGCTCTCGATCACACTTAGCTTTCCTCTTGCATGTCCTGTGTCAAGCAGCT
20060611C-008881 : TCTTCTTCGTCCACTCTTGTGAAACCTTCTGACCGGAGGCTGCTACAGCCTTGCCACTTC
---------+---------+---------+---------+---------+---------+ 278
SPL01_0053_F09.b : TCTTCTTCGTCCACTCTTGTGAAACCTTCTGACCGGAGGCTGCTACAGCCTTGCCACTTC
SPL01_0053_F08.b : TCTTCTTCGTCCACTCTTGTGAAACCTTCTGACCGGAGGCTGCTACAGCCTTGCCACTTC
20060611C-008881 : CTTTACGGGGCTCAAGCCCCCTCACACCCGTCATGCTCACTAGTACCCGATCATCCTCTG
---------+---------+---------+---------+---------+---------+ 338
SPL01_0053_F09.b : CTTTACGGGGCTCAAGCCCCCTCACACCCGTCATGCTCACTAGTACCCGATCATCCTCTG
SPL01_0053_F08.b : CTTTACGGGGCTCAAGCCCCCTCACACCCGTCATGCTCACTAGTACCCGATCATCCTCTG
20060611C-008881 : CCCGGGAGTGCCACCCCAGTTAACTGCCCAGGGATTCCGTCTCCACCAAAGAGCCTCTGC
---------+---------+---------+---------+---------+---------+ 398
SPL01_0053_F09.b : CCCGGGAGTGCCACCCCAGTTAACTGCCCAGGGATTCCGTCTCCACCAAAGAGCCTCTGC
SPL01_0053_F08.b : CCCGGGAGTGCCACCCCAGTTAACTGCCCAGGGATTCCGTCTCCACCAAAGAGCCTCTGC
20060611C-008881 : AAACCCTGCCTCCTGGAAAAACCTCTTCCCTATTGGTAATCCATGAATGCTAAGATGCAT
---------+---------+---------+---------+---------+---------+ 458
SPL01_0053_F09.b : AAACCCTGCCTCCTGGAAAAACCTCTTCCCTATTGGTAATCCATGAATGCTAAGATGCAT
SPL01_0053_F08.b : AAACCCTGCCTCCTGGAAAAACCTCTTCCCTATTGGTAATCCATGAATGCTAAGATGCAT
20060611C-008881 : TGTAGATATAATTTATATTCACGGCCATTTATGCCCTGCCTTATGTAGGCAGTTCAGCAC
---------+---------+---------+---------+---------+---------+ 518
SPL01_0053_F09.b : TGTAGATATAATTTATATTCACGGCCATTTATGCCCTGCCTTATGTAGGCAGTTCAGCAC
SPL01_0053_F08.b : TGTAGATATAATTTATATTCACGGCCATTTATGCCCTGCCTTATGTAGGCAGTTCAGCAC
20060611C-008881 : CAAGGAGAGTGCCTGTTTGCTTTTGCTTGGGTGCAAGGTCAGTGGAGGATTTACTGTGTT
---------+---------+---------+---------+---------+---------+ 578
SPL01_0053_F09.b : CAAGGAGAGTGCCTGTTTGCTTTTGCTTGGGTGCAAGGTCAGTGGAGGATTTACTGTGTT
SPL01_0053_F08.b : CAAGGAGAGTGCCTGTTTGCTTTTGCTTGGGTGCAAGGTCAGTGGAGGATTTACTGTGTT
20060611C-008881 : TAGGGCTCTGTAGTGCTGTTGGCACGATGTAATTTTTCCATAATAGATGAAAAGCCAAAC
---------+---------+---------+---------+---------+---------+ 638
SPL01_0053_F09.b : TAGGGCTCTGTAGTGCTGTTGGCACGATGTAATTTTTCCATAATAGATGAAAAGCCAAAC
SPL01_0053_F08.b : TAGGGCTCTGTAGTGCTGTTGGCACGATGTAATTTTTCCATAATAGATGAAAAGCCAAAC
20060611C-008881 : CCTGTATAAACACACTTTAATAATGGTGAAGGCACTGGCAGAGTTGTGTGCCGAGGAAAT
---------+---------+---------+---------+---------+---------+ 698
SPL01_0053_F09.b : CCTGTATAAACACACTTTAATAATGGTGAAGGCACTGGCAGAGTTGTGTGCCGAGGAAAT
SPL01_0053_F08.b : CCTGTATAAACACACTTTAATAATGGTGAAGGCACTGGCAGAGTTGTGTGCCGAGGAAAT
20060611C-008881 : ATTAAAGTTCACACTCCTGTAGCTTAGACTTAATTGAAGCCAGTAGTCTTCGTATTTACA
---------+---------+---------+---------+---------+---------+ 758
SPL01_0053_F09.b : ATTAAAGTTCACACTCCTGTAGCTTAGACTTAATTGAAGCCAGTAGTCTTCGTATTTACA
SPL01_0053_F08.b : ATTAAAGTTCACACTCCTGTAGCTTAGACTTAATTGAAGCCAGTAGTCTTCGTATTTACA
20060611C-008881 : TTTGGGTTGTTTGTTTGAATGTCAGACCTGGAGTGCTTCATCAGATGTCTTCCACTCCTC
---------+---------+---------+---------+---------+---------+ 818
SPL01_0053_F09.b : TTTGGGTTGTTTGTTTGAATGTCAGACCTGGAGTGCTTCATCAGATGTCTTCCACTCCTC
SPL01_0053_F08.b : TTTGGGGTGTTTGTTTGAATGTCAGACCTGGAGTGCTTCATCAGATGTCTTCCACTCCTC
20060611C-008881 : TACGTATAACTGCTTTAACCACCTG...................................
---------+---------+---------+---------+---------+---------+ 843
SPL01_0053_F09.b : TACGTATAACTGCTTaaacacctgccccaggattccctttggtctctatcttggctcccg
SPL01_0053_F08.b : TACGTATAACTGCTTTAACCACCTGcgnccaggattcccctttggtctctatcttgtctc
20060611C-008881 : ............................................................
---------+---------+---------+---------+---------+---------+ 843
SPL01_0053_F09.b : gcccagggaccgagtcgcattttcccttgtgtgccctggacatcctgacatggtgcctgg
SPL01_0053_F08.b : cgggcccagggacagaagtcgcatttttcttttgtgtgccttggacttccctgacatggt
20060611C-008881 : ............................................................
---------+---------+---------+---------+---------+---------+ 843
SPL01_0053_F09.b : cttcctttgctccaattatctctagggctgagaggcacagattcctcattacctgttgcc
SPL01_0053_F08.b : gcctggccttcccttgctccgcattaactccaaggctgaaaaggaaaaaattcctcatta
20060611C-008881 : ............................................................
---------+---------+---------+---------+---------+---------+ 843
SPL01_0053_F09.b : ttcctgacccaaaaaaaaaaaaaaaaaaaaggccccatgtgccc
SPL01_0053_F08.b : actgtttgcttccctgaccaaaaaaaaaaaaaaaaaaaaaaaggccccactgtgcctcaa
20060611C-008881 : ............................................................
---------+---------+---------+---------+---------+---------+ 843
SPL01_0053_F09.b :
SPL01_0053_F08.b : acctgcaagtc

![]()

